Acoplamiento molecular de derivados de la 6-(piridin)-7H-indeno[2,1-c]quinolina como inhibidores de la Topoisomerasa 1 y PARP 1
DOI:
https://doi.org/10.24054/bistua.v20i2.1434Keywords:
interacciones molecularesAbstract
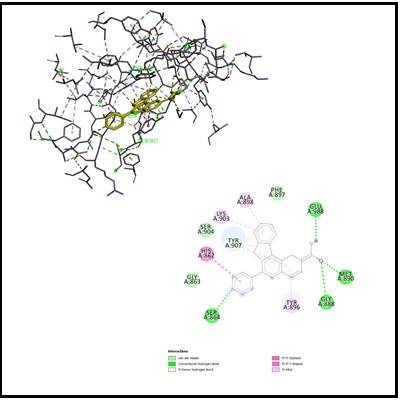
A variety of compounds from 7H-indeno[2,1-c]quinoline derivatives have been reported to possess substantial anticancer properties. However, their modes of action have not been clearly defined. Selected thirty 6-(pyridin)-7H-indeno[2,1-c]quinoline derivatives that exhibit anticancer activity were subjected to docking simulations using AutoDock Vina. To preliminarily investigate the potential molecular targets and to confirm the experimental activity testing for these anticancer compounds, the docking was performed using two enzymes involved with cell cycle, and DNA replication, i.e., topoisomerase and 1 and PARP 1 (Poly [ADP-ribose] polymerase 1). The docking results revealed that 6-(pyridin-4-yl)-7H-indeno[2,1-c]quinoline-2-carboxamide 1j exhibited better binding interaction to (Poly [ADP-ribose] polymerase 1) than the known D7N co-cristalized inhibitor. By other hand 2-fluoro-6-(pyridin-2-yl)-7H-indeno[2,1-c]quinoline 3e was best bound to DNA topoisomerase 1. The obtained results are useful to understand the structural features required to enhance the inhibitory activities.
Downloads
References
Li, J., Fu, A., & Zhang, L. (2019). An Overview of Scoring Functions Used for Protein–Ligand Interactions in Molecular Docking. Interdisciplinary Sciences – Computational Life Sciences, 11(2), 320–328. https://doi.org/10.1007/s12539-019-00327-w
Fu, Y., Zhao, J., & Chen, Z. (2018). Insights into the Molecular Mechanisms of Protein-Ligand Interactions by Molecular Docking and Molecular Dynamics Simulation: A Case of Oligopeptide Binding Protein. Computational and Mathematical Methods in Medicine, 2018. https://doi.org/10.1155/2018/3502514
Wu, Q., Peng, Z., Zhang, Y., & Yang, J. (2018). COACH-D: Improved protein-ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Research, 46(W1), W438–W442. https://doi.org/10.1093/nar/gky439
Ammar, Y. A., Sh El-Sharief, A. M., Belal, A., Abbas, S. Y., Mohamed, Y. A., Mehany, A. B. M., & Ragab, A. (2018). Design, synthesis, antiproliferative activity, molecular docking and cell cycle analysis of some novel (morpholinosulfonyl) isatins with potential EGFR inhibitory activity. European Journal of Medicinal Chemistry, 156, 918–932. https://doi.org/10.1016/j.ejmech.2018.06.061
AboulWafa, O. M., Daabees, H. M. G., & Badawi, W. A. (2020). 2-Anilinopyrimidine derivatives: Design, synthesis, in vitro anti-proliferative activity, EGFR and ARO inhibitory activity, cell cycle analysis and molecular docking study. Bioorganic Chemistry, 99(March), 103798. https://doi.org/10.1016/j.bioorg.2020.103798
Gurung, A. B., Ali, M. A., Bhattacharjee, A., Abul Farah, M., Al-Hemaid, F., Abou-Tarboush, F. M., Al-Anazi, K. M., Al-Anazi, F. S. M., & Lee, J. (2016). Molecular docking of the anticancer bioactive compound proceraside with macromolecules involved in the cell cycle and DNA replication. Genetics and Molecular Research, 15(2), 1–8. https://doi.org/10.4238/gmr.15027829
Fielden, J., Wiseman, K., Torrecilla, I., Li, S., Hume, S., Chiang, S. C., Ruggiano, A., Narayan Singh, A., Freire, R., Hassanieh, S., Domingo, E., Vendrell, I., Fischer, R., Kessler, B. M., Maughan, T. S., El-Khamisy, S. F., & Ramadan, K. (2020). TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts. Nature Communications, 11(1), 1–16. https://doi.org/10.1038/s41467-020-15000-w
Zhao, B., Liu, P., Fukumoto, T., Nacarelli, T., Fatkhutdinov, N., Wu, S., Lin, J., Aird, K. M., Tang, H. Y., Liu, Q., Speicher, D. W., & Zhang, R. (2020). Topoisomerase 1 cleavage complex enables pattern recognition and inflammation during senescence. Nature Communications, 11(1). https://doi.org/10.1038/s41467-020-14652-y
Takahashi, D. T., Gadelle, D., Agama, K., Kiselev, E., Zhang, H., Yab, E., Petrella, S., Forterre, P., Pommier, Y., & Mayer, C. (2022). Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states. Nature Communications, 13(1). https://doi.org/10.1038/s41467-021-27686-7
Additional Files
Published
Issue
Section
License
Copyright (c) 2022 © Autores; Licencia Universidad de Pamplona.

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
© Autores; Licencia Universidad de Pamplona





