Molecular compatibility analysis between proteins and their inhibitors: a case study in the formulation of the monoclonal antibody 4trp
Análisis de la compatibilidad molecular entre proteínas y sus inhibidores: un estudio de caso en la formulación del anticuerpo monoclonal 4trp
DOI:
https://doi.org/10.24054/bistua.v23i1.3563Keywords:
Potential energy, chemical instability, physical instability, mutation, isoelectric potentialAbstract
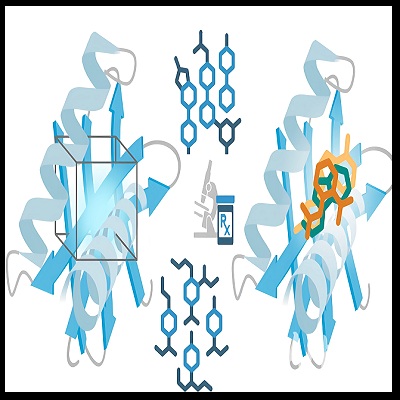
The incorporation of protease inhibitors (saquinavir, ritonavir, indinavir, nelfinavir, amprenavir, and lopinavir) into antiretroviral therapy has significantly reduced the morbidity and mortality associated with AIDS. These non-peptidic compounds potently and selectively inhibit HIV-1 protease. Therefore, an affinity study was conducted between the protease enzyme and each of its inhibitors by their binding energy using molecular docking to determine which inhibitor exhibits the highest binding affinity. Additionally, hydrogen bond interactions between the receptor and each ligand were analyzed to evaluate their correlation. From another perspective, this study examines the synthesis of the monoclonal antibody 4BINDING AFFINITY (IgG2a, derived from a murine IgG3 subclass antibody. Several mutations were introduced into the protein structure, replacing residues identified as high-risk for deamidation reactions. The analysis aimed to compare structural modifications to identify those that minimize aggregation risk. A quality assessment of the mutated product was conducted, supporting the stability improvements observed.
Downloads
References
Voshavar, C. (2019). Protease inhibitors for the treatment of HIV/AIDS: recent advances and future challenges. Current Topics in Medicinal Chemistry, 19(18), 1571-1598.
Ghosh, A. K., Weber, I. T., & Mitsuya, H. (2022). Beyond darunavir: Recent development of next generation HIV-1 protease inhibitors to combat drug resistance. Chemical Communications, 58(84), 11762-11782.
Singh, S., Tank, N. K., Dwiwedi, P., Charan, J., Kaur, R., Sidhu, P., & Chugh, V. K. (2018). Monoclonal antibodies: a review. Current clinical pharmacology, 13(2), 85-99.
Bayer, V. (2019, October). An overview of monoclonal antibodies. In Seminars in oncology nursing (Vol. 35, No. 5, p. 150927). WB Saunders.
Lee, W. Y. J., Fu, R. M., Liang, C., & Sloan, R. D. (2018). IFITM proteins inhibit HIV-1 protein synthesis. Scientific reports, 8(1), 14551.
York, J., Gowrishankar, K., Micklethwaite, K., Palmer, S., Cunningham, A. L., & Nasr, N. (2022). Evolving strategies to eliminate the CD4 T cells HIV viral reservoir via CAR T cell immunotherapy. Frontiers in Immunology, 13, 873701.
Antinori, A., Cicalini, S., Meschi, S., Bordoni, V., Lorenzini, P., Vergori, A., ... & HIV-VAC Study Group. (2022). Humoral and cellular immune response elicited by mRNA vaccination against SARS-CoV-2 in people living with HIV (PLWH) receiving antiretroviral therapy (ART) according with current CD4 T-lymphocyte count. Clinical Infectious Diseases.
Tolomeo, M., & Cascio, A. (2024). The complex dysregulations of CD4 T cell subtypes in HIV infection. International Journal of Molecular Sciences, 25(14), 7512.
Muñoz-Muela, E., Trujillo-Rodríguez, M., Serna-Gallego, A., Ruiz-Mateos, E., Espinosa, N., Roca-Oporto, C., ... & Gutiérrez-Valencia, A. (2022). Anti-CD4 autoantibodies in immunological nonresponder people with HIV: cause of CD4+ T-cell depletion?. AIDS, 36(9), 1207-1214.
Nkosi, T., Chasara, C., Papadopoulos, A. O., Nguni, T. L., Karim, F., Moosa, M. Y. S., ... & Ndhlovu, Z. M. (2022). Unsuppressed HIV infection impairs T cell responses to SARS-CoV-2 infection and abrogates T cell cross-recognition. Elife, 11, e78374.
Garcia, M. F. (2015). Farmacos Inhibidores de proteasas virales. Universidad Complutense de Madrid. Obtenido de http://147.96.70.122/Web/TFG/TFG/Memoria/MIGUEL%20FERNANDEZ%20GARCIA.pdf
Rose, Y., Duarte, J. M., Lowe, R., Segura, J., Bi, C., Bhikadiya, C., ... & Westbrook, J. D. (2021). RCSB Protein Data Bank: architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. Journal of molecular biology, 433(11), 166704.
Burley, S. K., Bhikadiya, C., Bi, C., Bittrich, S., Chen, L., Crichlow, G. V., ... & Zardecki, C. (2022). RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Science, 31(1), 187-208.
Additional Files
Published
Versions
- 2025-05-14 (2)
- 2025-05-12 (1)
Issue
Section
License
Copyright (c) 2025 © Autores; Licencia Universidad de Pamplona

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
© Autores; Licencia Universidad de Pamplona





